| |
Projects presently ongoing in our laboratory |
Aluminum Control of Organic Carbon Cycling in Temperate Forest
Ecosystems
The release of dissolved organic matter (DOM) from forest floor material
constitutes a significant flux of C to the mineral soil in temperate
forest ecosystems, with estimates on the order of 120 to 500 kg C
ha-1 year-1. Interaction of DOM with minerals and metals results
in sorptive fractionation and stabilization of OM within the soil
profile. In particular, aluminum (Al), which derives from crustal
rock, has been implicated recently as exerting significant control
over soil C cycling in a range of ecosystem types, but the underlying
mechanisms of stabilization - which involve tight coupling among
chemical, mineralogical and microbiological processes - are not well
known. We hypothesize that Al-DOM complexation impacts the
physical and chemical properties of DOM and, in turn, its sorption
and biodegradation. Further, we postulate that in a given
ecosystem type (and therefore with climatic variables fairly constant),
differences in C storage are controlled largely by the type of parent
rock.
Pinus ponderosa forests, established on basalt, granite and limestone
parent materials, are being used as model ecosystems for the proposed
work. Characterization of soil mineral assemblage, litter quality,
soil C MRT, and Al-tolerant microbes is being coupled with lab-based
experiments on soils collected from the sites to quantify Al-DOM
complexation, adsorption and biodegradation. Shifts in microbial
community with increasing Al-DOM complexation will be quantified
via DNA analysis of the microbes degrading 13C labeled Al-DOM complexes
in soil microcosms. |
| |
|
| |
Can soil genomics predict the impact of precipitation on nitrous
oxide (N2O) fluxes from soil?
Nitrous oxide (N2O) is a potent greenhouse gas and presently emissions
from soils are so poorly understood we can not predict how changes
in precipitation will impact the release of this important greenhouse
gas. Most nitrous oxide is released through biological processes,
especially nitrification and denitrification. These processes
not only affect the release of greenhouse gasses to the atmosphere
but also impact important ecosystem services such as purifying air
or water and regenerating soil fertility. Soil genomics, the
analysis of nucleic acids from soil, can provide the basis for new
ecosystem models by identifying marker sequences that estimate the
impact of our energy use on N2O fluxes from soil.
We have been studying four different ecosystems: mixed conifer
forest, ponderosa pine forest, pinyon-juniper biome, and grassland. The
sites have been subjected to three different precipitation treatments:
ambient, increased (+50%), and decreased (-30%), affected by redirecting
rainfall into the lysimeters that receive extra precipitation, and
by diverting rain from those in the low rainfall treatments. Preliminary
data show that adding moisture results in increased N2O fluxes
from mixed conifer and ponderosa pine soils but not the soils at
lower elevations. By analyzing the abundance and identity of genes
that encode enzymes critical in nitrification and denitrification
pathways we propose to identify marker sequences that predict the
impact of precipitation on nitrous oxide release. 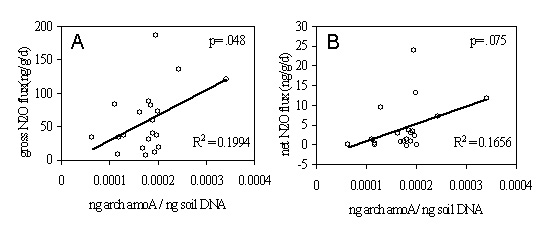
Comparison of archaeal amoA gene abundance with gross N2O fluxes (A)
and net N2O
fluxes (B) from mixed conifer soil. |
| |
|
| |
15N Natural Abundance of Soil Microbial Biomass as a Tool for
Assessing Controls on N-cycling Processes in Ecosystems
Nitrogen limits plant growth in many soils around the world. Plants
compete with microorganisms in the soil for nitrogen. When there is
lot of carbon, in the form of organic matter or dead organisms, and relatively
little nitrogen the microorganisms will take up most of the available nitrogen. In
contrast, when microbial growth is limited by carbon, soil microorganisms will
increase the amount of nitrogen available to plants by breaking down organic
matter that contains nitrogen. At present it is difficult to measure
if soil microorganisms are competing with plants for nitrogen or if they are
helping plants to grow by supplying nitrogen.
Nitrogen atoms occur in a least two forms in the environment. Most
of the atoms have 7 protons and 7 neutrons (14N) but a few atoms
will consist of 7 protons and 8 neutrons (15N). We have found
that soil organisms are enriched in 15N relative to nitrogen in the
soil solution and propose that the ratio of 15N to 14N in the microbial
biomass indicates if microbial growth is limited by nitrogen or by
carbon and that therefore it is possible to ascertain if plant growth
is promoted or restricted by soil organisms.
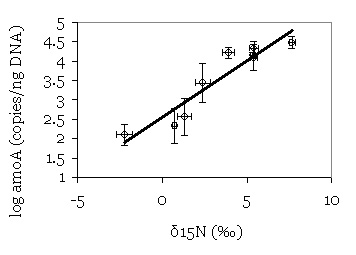
Relationship between natural abundance δ15N of DNA
extracted
from soil and abundance of archaeal amoA genes in soil. |
| |
|
| |
Ammonia oxidizing archaea in semi arid soils
Autotrophic ammonia-oxidizing communities, which are responsible
for the rate-limiting step of nitrification in most soils, have
not been studied extensively in semi-arid soils. We have been
measuring abundances of soil archaeal and bacterial amoA with
real-time PCR in semi-arid ecosystems along an elevation gradient
in northern Arizona. Archaeal amoA is the predominant form of
amoA at all sites, however archaeal amoA to bacterial amoA ratios
range from 17 to more than 1600. Though size of ammonia-oxidizing
bacteria populations are correlated with precipitation, temperature,
percent sand and soil C:N; we have not uncovered significant
relationships between ammonia-oxidizing archaea populations and
any of the environmental parameters evaluated in our studies.
Our results suggest that in these soils archaea may be the primary
ammonia-oxidizers, and that ammonia-oxidizing archaea and ammonia-oxidizing
bacteria occupy different niches. 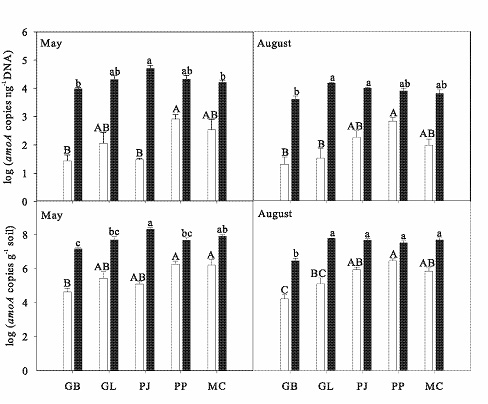
Abundance of bacterial (open bars) and archaeal (closed bars) amoA
genes
in soils from five different northern Arizonan ecosystems during
two different
seasons. Error bars show standard errors of the mean
and letters denote
significant differences among amoA abundances as
determined through Tukey
analysis (α = 0.05). |
| |
|
| |
Characterization of growing microorganisms in soil through stable
isotope probing with H218O.
We developed a new approach to characterize growing microorganisms
in environmental samples based on labeling microbial DNA with H218O. To
test if sufficient amounts of 18O could be incorporated into DNA
to use water as a labeling substrate for stable isotope probing (SIP),
Escherichia coli DNA was labeled by cultivating the bacteria in Luria
broth with H218O and labeled DNA was separated from [16O]DNA on a
cesium chloride gradient. Soil samples were incubated
with H218O for 6, 14 or 21 days and isopycnic centrifugation of the
soil DNA showed the formation of two bands after 6 days and three
bands after 14 or 21 days indicating that 18O can be used in stable
isotope probing of soil samples. Labeling soil DNA with H218O
allows identification of newly grown cells. In addition, cells
that survived but did not divide during an incubation period can
also be characterized with this new technique because their DNA will
remain without label. 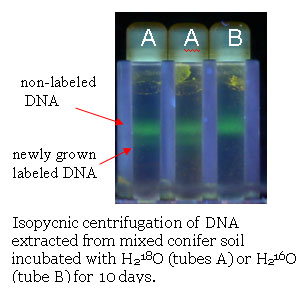 |
| |
|
| |
Biodiesel production from micro algae
Among the most important challenge facing the Unites States in the
21st century is the procurement of a dependable, environmentally
friendly source of energy. Present energy supplies are
dependent on fossil fuels, especially oil, which are predominantly
located in regions outside the United States and which upon incineration
release carbon dioxide, a greenhouse gas. Certain microalgae,
such as the diatom Cyclotella cryptica convert up to 50% of their
biomass into lipids which may, in turn, be transformed to biodiesel
through a simple chemical process. Biodiesel can be used
to power diesel engines in cars or to fuel mobile diesel powered
electricity generators. We have been studying Cyclotella cryptica
and have recovered a strain that can grow rapidly on a cheap
medium we have formulated. In addition we discovered that
lipid accumulation by this strain is strongly impacted by the
salinity of the medium.
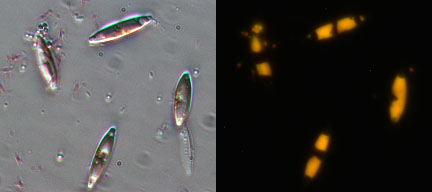
A lipid accumulating diatom isolated from Rio de Flag wetland in Flagstaff
AZ.
Light microscopy image of isolate stained with the dye Nile Red
is shown in A.
In panel B a fluorescence microscopy image of the same
sample is shown. In
a non-polar environment Nile Red fluoresces yellow
indicating the diatoms are
filled with lipids. |
| |
|
| |
Effects of Cattle Grazing on Relative Abundance of Soil Fungi and
Bacteria
The purpose of this study is to gain knowledge about grazing impacts
on microbial soil ecology north of the San Francisco Peaks and contribute
to the IGERT theme of “genes to environment.” Hypothesis
1: The ratio of fungi to bacteria found in soil will differ in the
site that is openly grazed versus the ones that are not openly grazed.
Hypothesis 2: A ratio of fungi/bacteria will show that more bacteria
are present in the grazed site and more fungi are present in the un-grazed
sites.
One gram of soil may contain millions of microorganisms, and
less than 1% of them can be cultured and viewed under microscopes.
So in order to obtain an accurate fungi/bacterial ratio, DNA will be
extracted from the soil core samples and real time polymerase chain
reaction (PCR) will be used to quantify relative gene abundance for
both bacteria and fungi. Using real time PCR is necessary to account
for all microorganisms. Two genes will be targeted by primers in this
study, one which is unique to nearly all bacteria, and one which is
unique to nearly all fungi. Other data to be
gathered in the study includes: soil moisture and soil nitrogen.
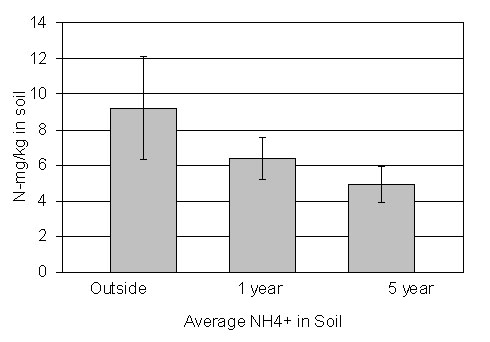
The openly grazed grassland site contains more NH4+ than
sites that are not grazed
for 1 year or not grazed for 5 years. |
| |
|
| |
Adaptation and Implementation of a water quality curriculum
in BIO 369 Environmental Microbiology
One of the most important environmental issues facing the Southwestern
US is the availability and quality of water. Water treatment
is therefore an increasingly important technology at the forefront
of environmental microbiology. We will adapt and implement
a new water quality curriculum, developed with funds from Arizona
state proposition 301, in an environmental microbiology course to
better prepare undergraduate microbiology students to face this challenge.
In addition, we will engage high school and community college students
from around Northern Arizona in water quality laboratory exercises.
The field of environmental microbiology has exploded in recent years,
merging molecular with traditional approaches to address the complex
roles of microorganisms in serious environmental problems. Rigorous
undergraduate training in this area requires hands-on experience
with multiple approaches to address real-world issues. The purpose
of the laboratory course in environmental microbiology at Northern
Arizona University (NAU) is to expose students to various aspects
of this field in order to acquaint them with key questions and methodologies
currently driving the field.
We have implemented a laboratory curriculum focused on water
quality microbiology by linking field experiences at the nearby Rio
De Flag Water Reclamation Facility with current molecular and functional
laboratory approaches. Students will acquire treated and untreated
water samples from this facility for determination of abundance of
viruses and organic pollutants. Student teams will also design,
conduct, and present experiments focused on the removal of viruses
or pollutants using samples from the reclamation facility.

Students from Shonto Preparatory Technical School
visiting NAU and
using electrophoresis equipment
purchased with funding from an NSF-CCLI
grant.
|
| |
|
|
|

