Introduction
Computing a distance matrix or the distance between all pairs of points is required in many applications. For example, direct n-body simulation requires computing the distance between all pairs of particles. In machine learning applications, such as k-nearest neighbor searches, hierarchical clustering, and density-based clustering require computing the distances between points in a dataset. Likewise, database applications, such as distance similarity searches, and range queries need to compute the distances between (potentially all) points in a dataset. Given the range of applications that require computing a distance matrix, this module explores computing a distance matrix in parallel using MPI.
Student Learning Outcomes
- Understand performance and scalability of compute-bound algorithms.
- Understand how cache reuse can be exploited to improve performance through the use of tiling.
- Understand the performance trade-off between small and large tile sizes.
- Utilize a performance tool to measure cache misses.
Prerequisites
Some knowledge of the MPI API and familiarity with the SLURM batch scheduler.
MPI Primitives Used
- MPI_Scatter
- MPI_Reduce
Files Required
Problem Description
As described above, a distance matrix is useful in many contexts. The problem is outlined as follows, for each point in a dataset D, compute the distance between it and every other point. The distance between each point is stored in an N×N distance matrix (implemented as a 2-D array). The problem has a time complexity of O(N2), where there are N points in the dataset. Use the Euclidean distance to compute the distance between points. The Euclidean distance between point a∈D and b∈D is outlined as follows: dist(a,b)=√∑dj=1(xaj−xbj)2, where each point pi∈D has coordinates in d dimensions, and each coordinate is denoted as xj, where j=1,…,d.
To get started, you will be provided with a file distance_matrix_starter.c. The program will import a dataset, and expects the following command line arguments: <N> <DIMS> <BLOCKSIZE> <FNAME>, where:
- N is the number of data points.
- DIMS is the data dimensionality.
- BLOCKSIZE is the block size when tiling the computation in programming activity #2.
- FNAME is the dataset filename.
We will use a real-world dataset, the Million Song Dataset. We will not use the entire dataset, rather we will select the first N=100,000 rows of the original dataset. The dataset has 90 features (d=90).
- Q1: Assume the dataset is stored as double precision floating point values in main memory (each double requires 8 bytes of space). How much memory (in MiB) is required to store the entire dataset in main memory?
- Q2: Assume the distance matrix is stored using double precision floating point values in main memory (each double requires 8 bytes of space). How much memory (in MiB) is required to store the entire distance matrix in main memory?
- Q3: Could you store the dataset in main memory on a typical laptop computer? Explain.
- Q4: Could you store the distance matrix in main memory on a typical laptop computer? Explain.
Computing the Distance Matrix
Figure 1 outlines the storage of the distance matrix. Note the following:
- The distance matrix is stored as a 2-D array.
- You will assign N/p points/rows to compute by each process rank, where there are p total process ranks. Thus, each rank will compute the distance between its assigned points and every other point in the dataset, D. If N mod p !=0, you must assign the “leftover” rows/points to the other ranks. You may assign all leftover points to the last rank, since the number of leftovers will be small.
- You may replicate the dataset across all process ranks, as each rank requires the data of all other points in the dataset. The starter file will import the dataset file on all ranks.
- The distance matrix must be left distributed at the end of the computation. Memory should only be allocated for the local distance matrix stored on each rank.

Figure 1: Example of computing and storing the distance matrix with N=12 points on p=4 ranks, where each rank is assigned N/p=3 points. Rank 0, 1, 2, 3 compute the red, blue, yellow, and green rows, respectively.
- Q5: When using p=1 and p=20 ranks, what is the total memory required (in MiB) to store the distance matrix, respectively?
Example Input and Output
Consider the following dataset with N=4 and d=4.
Input Dataset:
1.0,2.0,3.0,4.0
6.0,7.0,8.0,9.0
11.0,12.0,13.0,14.0
15.0,16.0,17.0,18.0
Output Distance Matrix:
0.000000, 10.000000, 20.000000, 28.000000,
10.000000, 0.000000, 10.000000, 18.000000,
20.000000, 10.000000, 0.000000, 8.000000,
28.000000, 18.000000, 8.000000, 0.000000,
How to Time Sections of a Program
In this module you will time a section of the program. The code below shows an example of timing a section of code.
double tstart=MPI_Wtime();
//code to time
double tend=MPI_Wtime();
printf("Time (s): %f", tend - tstart);
If you want to time your code when all ranks in a communicator have reached a certain point in the program, you can use a barrier as follows (the example below shows synchronization across the MPI_COMM_WORLD communicator):
MPI_Barrier(MPI_COMM_WORLD);
double tstart=MPI_Wtime();
//code to time
double tend=MPI_Wtime();
printf("Time (s): %f", tend - tstart);
Programming Activity #1
Modify distance_matrix_starter.c and implement a solution to compute the distance matrix.
In your solution, compute the distance between each point and every other point in the row. For example, in Figure 2, Rank 0 computes the distance between the point p0 and p0, p1, …, p11, then it computes the distance between point p1 and p0, p1, …, p11, and so on. This is a baseline approach to computing the distance matrix – iterating over the rows and columns requires two nested for loops.
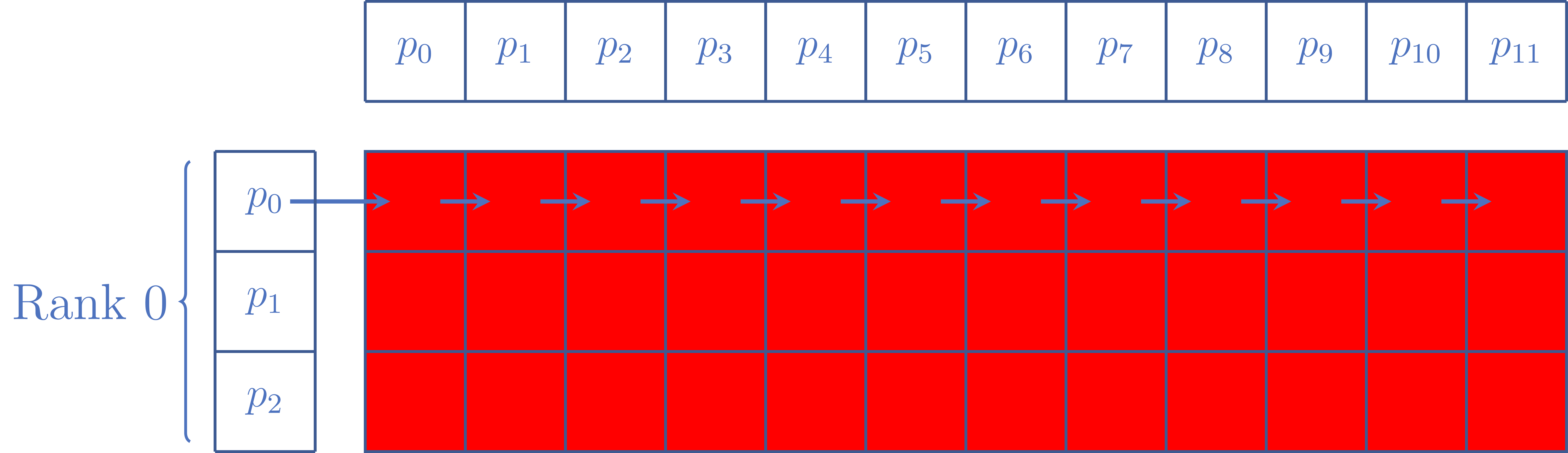
Figure 2: Illustrative example of the order of calculations. Compare each row/point to all other points before computing the next row.
In your solution, you must perform the following tasks:
- Have rank 0 evenly split the work as ranges of rows that are assigned to each process rank. You must use
MPI_Scatter()to send the row information to the process ranks. - To validate that your solution is correct, you must output the distance matrix and perform a
diffbetween a sequential reference implementation that you have validated works correctly and your parallel implementation on a small distance matrix. - Once you have validated your parallel implementation for a few cases, you must check that your solution is correct by simply adding up the sums of all distances computed locally by each rank, and then perform a reduction using
MPI_Reduce()at rank 0 that adds up the global sum of all distances in the distance matrix. You will report the global sums in your report.
When running your program on the cluster, make sure your job script(s) and program conforms to the following guidelines:
- All programs are compiled with the
-O3flag. For example:mpicc -O3 distance_matrix_starter.c -lm -o distance_matrix_starter. - To estimate the memory usage required of your job, you can use your responses to the questions above.
- Use a single node.
- Use the exclusive flag for all experiments so that your results are not polluted by other users running on the node.
- Run all of your experiments on a single node generation (if applicable).
- You will time the execution of your program using
MPI_Wtime(). Begin timing after you have imported the dataset, and have allocated memory for the distance matrix after all of the ranks have been sent their respective ranges. Since the workloads are evenly distributed between process ranks, it is sufficient in this exercise to compute the response time on rank 0 only (instead of computing the total response time using the longest executing process rank). - Do not include in your time measurements the time to compute the global sum.
- Run three time trials for each execution configuration. Report the average response time in seconds (s). All other metrics you will report are derived from the response time.
- You can enter any value for the block size on the command line. This block size is used in Programming Activity #2.
In your report, include a Table with the following information for executions with 1, 4, 8, 12, 16, and 20 process ranks. The report must include the response time, the parallel speedup, the parallel efficiency, the global sum used for validation, and the job script name that you used to obtain these results. The job script may be the same for all executions. Include all job scripts in the submitted archive. A template table is provided below.
| # of Ranks (p) | Time (s) | Parallel Speedup | Parallel Efficiency | Global Sum | Job Script Name (*.sh) |
|---|---|---|---|---|---|
| 1 | |||||
| 4 | |||||
| 8 | |||||
| 12 | |||||
| 16 | |||||
| 20 |
Validation
The global sum should be close to 455386000.680 (there will be floating point arithmetic error).
Answer the following questions:
- Q6: Do you think the performance of the distance matrix calculation is good? Explain.